Rows: 3,818
Columns: 13
$ name <chr> "Alabama A & M University", "University of Alabama at …
$ title_iv <chr> "Title IV postsecondary institution", "Title IV postse…
$ carnegie_class <chr> "Master's Colleges & Universities: Larger Programs", "…
$ state <chr> "Alabama", "Alabama", "Alabama", "Alabama", "Alabama",…
$ total_enroll <dbl> 6007, 21639, 647, 9237, 3828, 38644, 1777, 2894, 5109,…
$ pct_admitted <dbl> 68, 87, NA, 78, 97, 80, NA, NA, 92, 44, 57, NA, NA, NA…
$ n_bach <dbl> 511, 2785, 54, 1624, 480, 6740, NA, 738, 672, 5653, 26…
$ n_mast <dbl> 249, 2512, 96, 570, 119, 2180, NA, 80, 300, 1415, 0, N…
$ n_doc <dbl> 9, 166, 20, 41, 2, 215, NA, 0, 0, 284, 0, NA, 0, NA, N…
$ tuition_fees <dbl> 10024, 8568, NA, 11488, 11068, 11620, 4930, NA, 8860, …
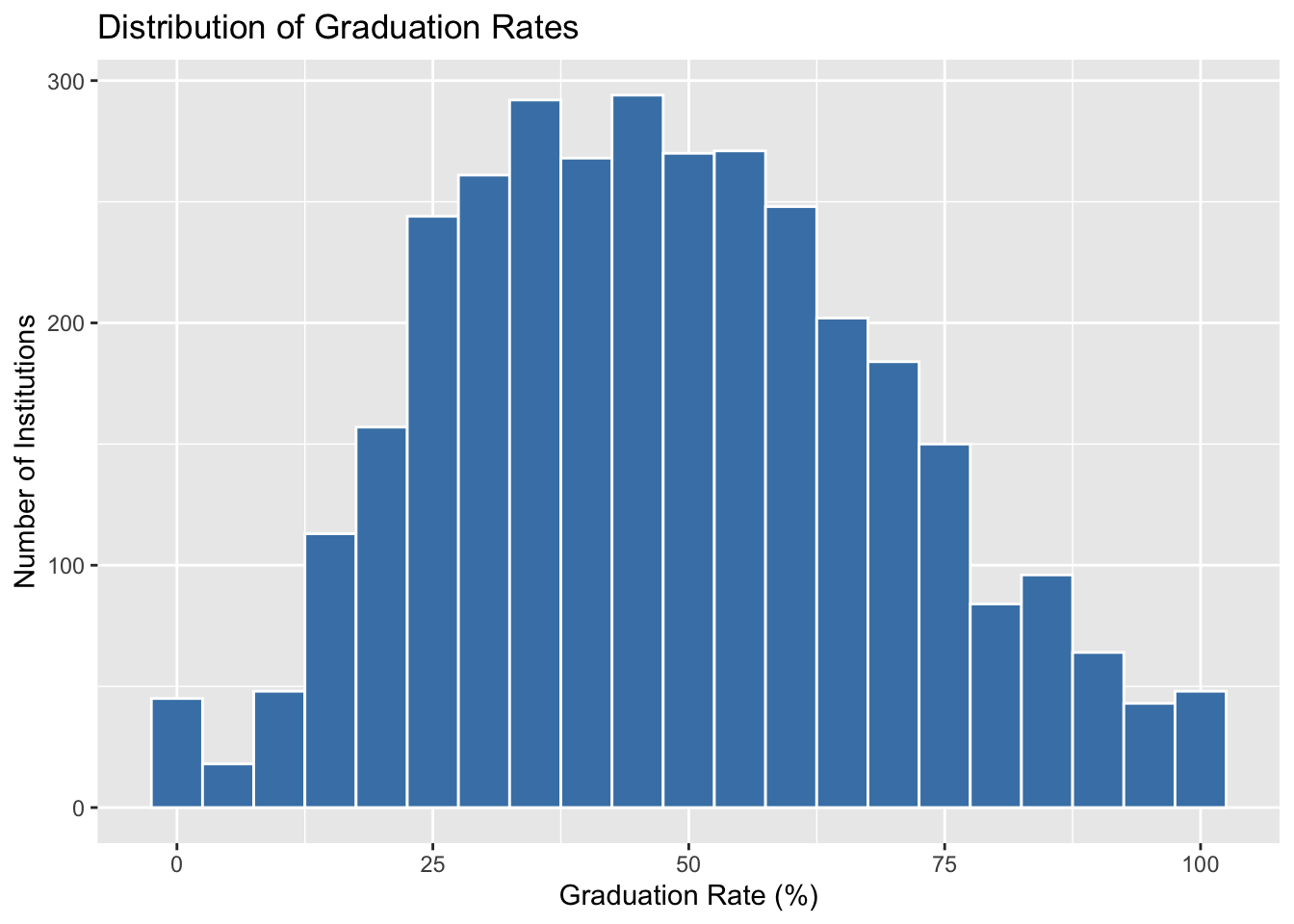
$ grad_rate <dbl> 27, 64, 50, 63, 28, 73, 22, NA, 36, 81, 65, 26, 9, 29,…
$ percent_fin_aid <dbl> 87, 96, NA, 96, 97, 87, 87, NA, 99, 79, 100, 96, 92, 8…
$ avg_salary <dbl> 77824, 106434, 36637, 92561, 72635, 97394, 63494, 8140…