class: clear, title-slide, inverse, center, top, middle # LASER Learning Lab Orientation ## Reproducible Research & The Data-Intensive Research Workflow ### The LASER Team ### July 17, 2023 --- # Agenda .pull-left[ ## Part 1: Core Concepts ### Reproducible Research with RStudio - What is reproducible research? - The benefits of reproducibility - The tools of reproducible research ] .pull-right[ ## Part 2: R Code-Along ### Data-Intensive Research Workflow - Prepare - Wrangle - Explore - Model - Communicate ] ??? Each Learning Lab begins with a 60-minute interactive presentation consisting of two parts. The first focuses on a conceptual overview of key terminology, techniques, and applications. The second part provides a short but highly structured code-along activity that demonstrates key packages and functions required for specific data analysis techniques highlighted in each unit and an exemplary research study. In this learning lab we'll focus on the importance of reproducible research and tools for reproducible research; and then gain some hands on experience with the data-intensive research workflow to conduct a simple analysis with R. Everything we introduce in this presentation you will have an opportunity to explore in much greater depth through the essential readings and a coding case study after the break. --- class: clear, inverse, middle, center # Part 1: Core Concepts ---- Reproducible Research with RStudio --- class: clear, laser .quote[“...future you and anyone else will be able to **pick up your analysis and reproduce the same results**, including figures and tables.”] .author[~ Harrison, 2020] --- # What is Reproducible Research? .pull-left[ In computational sciences like LA: - Ideally... researchers can ***replicate*** your findings by following the procedures used to gather the data and run the computer code. - Realistically... the data and code used to make a finding are available and sufficient to ***recreate*** the finding. ] .pull-right[ .center[<img src="img/reproducible-research.png" height="400px"/> ] ] .footnote[ Gandrud, C. (2013). [Reproducible research with R and R studio (3rd Edition).](https://github.com/christophergandrud/Rep-Res-Book). CRC Press. ] ??? The information from this conceptual overview on reproducible research is drawn largely from the excellent book by Christopher Gandrud,Reproducible research with R and R studio. The distinction between these two definitions is that we recognize it is sometimes difficult to replicate the original data set in a study because of issues such as limited resources to gather new data, or because the original study may have already sampled the full universe of cases. --- # The Benefits of Reproducible Research .pull-left[ **For Science:** - Standard to judge scientific claims - Enhances *replicability* - Avoiding effort duplication - Cumulative knowledge development ] .pull-right[ **For Yourself:** - Better work habits - Better teamwork - Changes are easier - Higher impact research ] ??? ##For Science ###Standard to judge scientific claims Reproducibility opens up your findings and work to scrutiny, and is the **minimum** standard for judging scientific claims. However, “a study can be reproducible but still be wrong” (Peng, 2014). ###Reproducibility enhances replicability. If other researchers are able to clearly understand how a finding was originally made, then they will be better able to conduct comparable research, in meaningful attempts to replicate the original findings. However, a finding in one study may be statistically significant when **reproduced** using the original data/code, but when researchers try to **replicate** it using new data and even methods, they are unable to find a similar result. The original finding could have been noise, even though it is fully reproducible. ### Avoiding effort duplication Reproducible research, in theory, should cut down on the amount of time others have to spend gathering data or developing procedures that have already been collected or figured out. ### Cumulative knowledge development They can instead use that time to more quickly build on established findings and develop new knowledge. ## For Yourself ### Better work habits It can spur you to more effectively plan and organize your research. It should push you to bring your data and source code up to a higher level of quality than you might if you thought no one was looking. ### Better teamwork RR makes it easier for your collaborators figure out what you have done also to understand your work and up to speed on a research project. ### Changes are easier No actual research process is completely linear. You almost never gather data, run analyses, and present your results without going backwards to add variables, make changes to your statistical models, create new graphs, alter results tables in light of new findings. . RR makes it easier to reproduce parts or even all of your own research. ### Higher impact research Reproducible research is more likely to be useful for other researchers than nonreproducible research. Useful research is cited more frequently! --- class: middle, no-logo # Reproducible Research? <img src="img/phd-comic.jpeg" height="500px"/> ??? In practice, your research should be easy for independent researchers to reproduce! If a study is difficult to reproduce, it’s more likely that no one will reproduce it. If someone does attempt to reproduce it, it will be difficult for them to tell if any errors they find were in the original research or problems introduced during the reproduction. So next we're going to talk about some general best practices for reproducible research, and then give an overview of the research tools we'll be working with throughout the LASER institute to help ensure it's relatively easy to reproduce our research. --- # Discussion Think about the following questions and then discuss at your table: - What have your experiences been with reproducible research? - What tools have you used to ensure the reproducibility of your work? - What questions do you have about reproducible research? --- # Best Practices for Reproducible Research 1. Document everything! 2. Everything is a (text) file. 3. All files should be human readable. 4. Explicitly tie your files together. 5. Have a plan to organize, store, and make your files available. To learn more: [Reproducible research with R and R studio (3rd Edition)](https://github.com/christophergandrud/Rep-Res-Book). CRC Press. ??? ## 1. Document everything! Obviously, in order to reproduce your research, others must be able to know what you did. Documentation of includes, but is not limited to the data itself, analysis code, the write-up of results, and explanations of these files (e.g. data set codebooks, software information, and so on). ## 2. Everything is a (text) file. Ideally, your documentation will be stored in the simplest file format possible to store this information. Usually the simplest file format is the humble, but versatile, text file. Text files are extremely nimble and can used your data in, like in comma-separated values (CSV) format for example, can contain your analysis code in files, and can be the basis for publishable formats like articles, webpages, books, etc. using markup languages such as Markdown. One reason reproducible research is best stored in text fles is that this helps future-proof your research ## 3. All files should be human readable. Treat all of your research fles as if someone who has not worked on the project will, in the future, try to understand them. Computer code is a way of communicating with the computer. It is ‘machine readable’ in that the computer is able to use it to understand what you want to do.2 However, there is a very good chance that other people (or you six months in the future) will not understand what you were telling the computer. So, you need to make all of your files ‘human readable’. To make them human readable, you should comment on your code with the goal of communicating its design and purpose. ## 4. Explicitly tie your files together. Researchers often do not explicitly document the relationships between fles that they used in their research. For example, the results of an analysis–a table or figure–may be copied and pasted into a presentation document. It can be very difficult for future researchers to trace the table or fgure back to a particular statistical model and a particular data set without clear documentation. Therefore, it is important to make the links between your files explicit. ## 5. Have a plan to organize, store, and make your files available. Finally, in order for independent researchers to reproduce your work, they need to be able access the fles that instruct them how to do this. Files also need to be organized so that independent researchers can fgure out how they ft together. So, from the beginning of your research process, you should have a plan for organizing your fles and a way to make them accessible. ## Learn More We've only scratched the surface Reproducible Research practices and highlight recommend the book Reproducible research with R and R studio to learn more. --- class: no-logo # The Tools of Reproducible Research .panelset[ .panel[.panel-name[Tool Types] Reproducible research involves two broad sets of tools: 1. *A Reproducible Research Environment*, that includes the statistical tools you need to run your analyses; automatically track the provenance of data, analyses, and results; and to package them for redistribution”. 2. *A Reproducible Research Publisher* the prepares dynamic documents for presenting results and is easily linked to the reproducible research environment. ] .panel[.panel-name[Posit Cloud] .center[ <img src="img/posit-cloud.png" height="350px"/> ] .center[[go.ncsu.edu/laser-labs](https://go.ncsu.edu/laser-labs) ] ] .panel[.panel-name[RStudio] .center[ <img src="img/rstudio.png" height="425px"/> ] ] .panel[.panel-name[R Markdown] .center[ <img src="img/r-markdown.png" height="425px"/> ] ] .panel[.panel-name[GitHub] .center[ <img src="img/github.png" height="350px"/> ] .center[[go.ncsu.edu/laser-github](https://go.ncsu.edu/laser-github) ] ] .panel[.panel-name[R] .center[ <img src="img/r-script.png" height="425px"/> ] ] ] ??? ## Tool Types N/A ## Posit Cloud Posit Cloud (formerly RStudio Cloud) provides a powerful set of data science tools right in your browser with no installation or complex configuration required. All of our Learning Lab materials will be access through Posit Cloud this year so we encourage you bookmark posit.cloud in your browser. < Have scholars navigate to our LASER Learning Labs 2023 workspace using the link provide and provide a breif overview of the the Posit Cloud features like workspaces, primers, cheatsheets, etc. > ## RStudio < Next have scholars navigate to our laser-orientation RStudio Project "assignment" and click the link to make a copy for themselves and open up the project. > RStudio is an integrated development environment (IDE) for R and Python and includes a console for running R code directly, syntax-highlighting editor that supports direct code execution in the source pane, and tools for plotting, history, debugging, and management of research projects in the Environment and Files panes. RStudio is basically your home for all of the files, images, reports, and code that are used in a research project given project. When you create a "project" with RStudio, it's straightforward to divide your work into multiple contexts, each with their own working directory, workspace, history, and source documents. < Provide a brief walkthrough of the different panes and discuss what they are used for including how they might make their own R Project is they so desired. > ## R Markdown < Show scholars how to navigate to the files tab and open up the file named laser-lab-slides.Rmd> As we noted earlier, plain text files written in markup languages like Markdown can be the basis for documents such as presentations, webpages, dashboards, and books. This R Markdown file can be opened not just with RStudio, but a simple text editor built into most operating systems. This specific text file is what was used to make the slides you're looking at now. Click the "knit" button located at the top of the toolbar to convert this markdown file into an HTML slide show. I should appear in the Viewer table or in a separate browser window and you will need it for our Part 2 code along. ## GitHub GitHub is an interface and cloud hosting service built on top of the Git version control system. GitHub was not explicitly designed to host research projects or even data, but for programmers to work together to build computer programs. This seems very far from reproducible research but remember that as reproducible researchers, we are building projects out of interconnected text fles. In important ways, this is exactly the same as building a computer program and offers many highly developed and robust features for reproducible researchers. We won't be using GitHub in our Learning Labs, but I did want to note that if you click the link below, it will take you to our GitHub organization site where you will find our RStudio Projects stored as Repositories, or repos, including the laser-orientation repository you just opened. ## R Underlying all of this is R, a programming language originally for statistics and graphics but has evolved into rich platform for data science and learning analytics that can be used to create a wide range of static and dynamic outputs for communicating your research. Let's create a new R script so we can begin working with R to help illustrate a simple research workflow and analyze some data. < Direct scholars to create a new R script as illustrated in the screenshot. Show them how to name the file and save to their main directory. > --- class: clear, inverse, middle, center # part_2(R, code_along) ---- The Data-Intensive Research Workflow --- # The Data-Intensive Research Workflow .center[ <img src="img/laser-cycle.png" height="400px"/> ] .footnote[ Krumm, A., Means, B., & Bienkowski, M. (2018). [Learning Analytics Goes to School.](https://www.routledge.com/Learning-Analytics-Goes-to-School-A-Collaborative-Approach-to-Improving/Krumm-Means-Bienkowski/p/book/9781138121836). Routledge. ] ??? --- class: no-logo # Prepare .panelset[ .panel[.panel-name[Research Context] .pull-left[ ## The Study The setting of this study was a public provider of individual online courses in a Midwestern state. Data was collected from two semesters of five online science courses and aimed to understand students motivation. ] .pull-right[ ## Research Question Is there a relationship between the time students spend in a learning management system and their final course grade? ] ] .panel[.panel-name[Packages] Sometimes referred to as "libraries", packages are shareable collections of R code that can contain functions, data, and/or documentation. ```r # Load the {tidyverse} package library(tidyverse) ``` ``` ## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ── ## ✔ dplyr 1.1.4 ✔ readr 2.1.5 ## ✔ forcats 1.0.0 ✔ stringr 1.5.1 ## ✔ ggplot2 3.5.0 ✔ tibble 3.2.1 ## ✔ lubridate 1.9.3 ✔ tidyr 1.3.1 ## ✔ purrr 1.0.2 ## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ── ## ✖ dplyr::filter() masks stats::filter() ## ✖ dplyr::lag() masks stats::lag() ## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors ``` ```r # How to install the package if not already installed # install.packages("tidyverse") # Note this code is commented out so it will not run ``` ] .panel[.panel-name["Read" Data] The {readr} tidyverse package has several functions like `read_csv()` for importing rectangular data from delimited text files such as comma-separated values (CSV), a preferred file format for reproducible research. ```r # "read" into your R environment the sci-online-classes.csv file # stored in the data folder of your R project and # save as a new data "object" named sci_data sci_data <- read_csv("data/sci-online-classes.csv", col_names = TRUE) ``` R has also packages for proprietary data formats like .xls, .sas, and .dat. ] .panel[.panel-name[Inspect Data] You should now see a new data "object" named `sci_data` saved in your Environment pane. Try clicking on it and see what happens! Now let's "print" the output to our Console to view another way: ```r # print data to console to view sci_data ``` What variables do you think might help us answer our research question? ] .panel[.panel-name[R Syntax] The R language can express complex ideas like in this simple "sentence": `sci_data <- read_csv("data/sci-online-classes.csv", col_names = TRUE)` - *Functions* are like verbs: `read_csv()` - *Objects* are the nouns: `read_csv("data/sci-online-classes.csv")` - *Arguments* are like adverbs: `read_csv("data/sci-online-classes.csv", col_names = TRUE)` - *Operators* are like “punctuation” `sci_data <- read_csv("data/sci-online-classes.csv", col_names = TRUE)` ] ] --- class: # Wrangle Data wrangling is the process of cleaning, ["tidying"](https://r4ds.had.co.nz/tidy-data.html?q=tidy%20data#tidy-data-1), and transforming data. In Learning Analytics, it often involves merging (or joining) data from multiple sources. Since we are interested the relationship between time spent in an online course and final grades, let's `select()` the `FinalGradeCEMS` and `TimeSpent` from `sci_data`. ```r # select student_id and course_id and save as sci_data |> select(FinalGradeCEMS, TimeSpent) ``` Note the use of a the powerful `|>` operator called a **pipe**, which are used for combining a sequence of functions or processes. --- class: no-logo # Explore .panelset[ .panel[.panel-name[EDA] Exploratory data analysis involves processes of **describing** your data numerically or graphically and often involves: - **calculating** summary statistics like frequency, means and standard deviations - **visualizing** your data through charts and graphs EDA can be used to help answer research questions or generate new questions about your data, discover relationships between and among variables, and create new variables (i.e., feature engineering) for data modeling. ] .panel[.panel-name[Graph Template] The [{ggplot2}](https://ggplot2.tidyverse.org) package follows a common [graphing workflow](https://r4ds.had.co.nz/data-visualisation.html?q=template#a-graphing-template) for making graphs. To make a graph, you simply: 1. Start the graph with `ggplot()` function and include your data as an argument; 2. "Add" elements to the graph using the `+` operator `a geom_()` function; 3. Select variables to graph on each axis with the `aes()` argument. A common graphing template is as simple as two lines of code: ```r ggplot(<DATA>) + <GEOM_FUNCTION>(aes(x = <VARIABLE1>, y = <VARIABLE2>)) ``` ] .panel[.panel-name[Our First Graph!] Scatterplots use the point **geom**, i.e., the `geom_point()` function, and are most useful for displaying the relationship between two continuous variables. ```r # make a scatter plot showing time spent and final grades ggplot(sci_data) + geom_point(aes(x = TimeSpent, y = FinalGradeCEMS)) ``` ] .panel[.panel-name[Interpret Graph] .pull-left[Hopefully your scatterplot looks like something like the one to the right. *How would you interpret this graph?* ] .pull-right[ 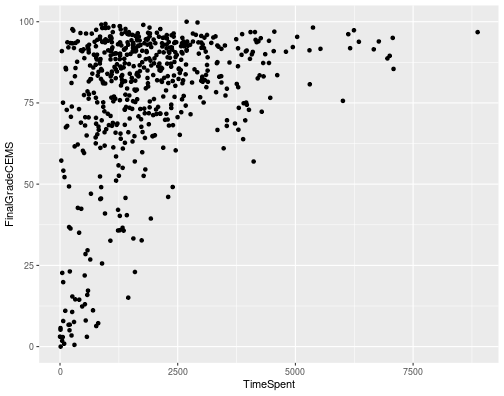<!-- --> ] ] ] --- class: no-logo # Model .panelset[ .panel[.panel-name[Goal] Modeling involves a simple low-dimensional summary of a dataset. According to Krumm et al. (2018), there are two general types of modeling approaches: - **Unsupervised** learning algorithms can be used to understand the structure of one’s dataset. - **Supervised** models, on the other hand, help to quantify relationships between features and a known outcome. Ideally, a good model will separate true “signals” (i.e. patterns generated by the phenomenon of interest) from the “noise” (i.e. random variation that you’re not interested in). ] .panel[.panel-name[A Simple Model] We'll dive much deeper into modeling in subsequent learning labs, but for now let's see if there is a statistically significant relationship between students' final grades, `FinaGradeCEMS`, and the `TimeSpent` in the LMS: ```r # use a simple linear regression model to predict final course grades m1 <- lm(FinalGradeCEMS ~ TimeSpent, data = sci_data) summary(m1) ``` ] .panel[.panel-name[Interpret] .code70.remark-code[ ``` ## ## Call: ## lm(formula = FinalGradeCEMS ~ TimeSpent, data = sci_data) ## ## Residuals: ## Min 1Q Median 3Q Max ## -67.136 -7.805 4.723 14.471 30.317 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 6.581e+01 1.491e+00 44.13 <2e-16 *** ## TimeSpent 6.081e-03 6.482e-04 9.38 <2e-16 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 20.71 on 571 degrees of freedom ## (30 observations deleted due to missingness) ## Multiple R-squared: 0.1335, Adjusted R-squared: 0.132 ## F-statistic: 87.99 on 1 and 571 DF, p-value: < 2.2e-16 ``` ] ] ] --- class: no-logo # Communicate .panelset[ .panel[.panel-name[Data Products] Krumm et al. (2018) have outlined the following 3-step process for communicating finding with education stakeholders: 1. **Select.** Selecting analyses that are most important and useful to an intended audience, as well as selecting a format for displaying that info (e.g. chart, table). 2. **Polish.** Refining or polishing data products, by adding or editing titles, labels, and notations and by working with colors and shapes to highlight key points. 3. **Narrate.** Writing a narrative pairing a data product with its related research question and describing how best to interpret and use the data product. ] .panel[.panel-name[Dashboards] .center[ <img src="img/dashboards.png" height="350px"/> ] .center[[https://pkgs.rstudio.com/flexdashboard](https://pkgs.rstudio.com/flexdashboard/)] ] .panel[.panel-name[Websites] .center[ <img src="img/websites.png" height="350px"/> ] .center[[https://pkgs.rstudio.com/blogdown/](https://pkgs.rstudio.com/blogdown/)] ] .panel[.panel-name[Books] .center[ <img src="img/books.png" height="350px"/> ] .center[[https://pkgs.rstudio.com/bookdown/](https://pkgs.rstudio.com/bookdown/)] ] .panel[.panel-name[And More] R Markdown supports dozens of static and dynamic output formats including [HTML](https://bookdown.org/yihui/rmarkdown/html-document.html), [PDF](https://bookdown.org/yihui/rmarkdown/pdf-document.html), [MS Word](https://bookdown.org/yihui/rmarkdown/word-document.html), [Beamer](https://bookdown.org/yihui/rmarkdown/beamer-presentation.html), [HTML5 slides](https://bookdown.org/yihui/rmarkdown/ioslides-presentation.html), [Tufte-style handouts](https://bookdown.org/yihui/rmarkdown/tufte-handouts.html), [books](https://bookdown.org), [dashboards](flexdashboard/), [shiny applications](https://bookdown.org/yihui/rmarkdown/shiny-documents.html), [scientific articles](https://github.com/rstudio/rticles), [websites](https://bookdown.org/yihui/rmarkdown/rmarkdown-site.html), and more. ] ] ??? --- # What's Next? .pull-left[ ## Your First LASER Badge! Next you will complete an interactive "case study" which is a key component to each learning lab. Navigate to the Files tab and open the following file: `laser-lab-case-study.RMD` ] .pull-right[ ## Essential Readings - [Reproducible Research with R and RStudio](https://github.com/christophergandrud/Rep-Res-Book) (chapters 1 & 2) - [Learning Analytics Goes to School](https://www.routledge.com/Learning-Analytics-Goes-to-School-A-Collaborative-Approach-to-Improving/Krumm-Means-Bienkowski/p/book/9781138121836) (pages 28 - 58) - [Data Science in Education Using R](https://datascienceineducation.com) - [R for Data Science](https://r4ds.had.co.nz/index.html) ] --- class: clear, center ## .font130[.center[**Thank you!**]] <br> <img src="img/laser-instructors.png" height="350px"/>